Spotlight Program
Showcasing outstanding research by Swiss life scientists.
Research in the spotlight

Elena Foli & Mehrpouya Mostanfar
In this study, Niccolò, Elena, Mehrpouya, and their collaborators functionally characterized the FCRL3 immune receptor and unraveled its role in memory T cell regulation.
Read more

Ilaria Fregno & Mikhail Rudinskiy
Together with their colleagues, Ilaria and Mikhail developed and characterized the mode of action of two structurally targeted allosteric regulators (STARs) of GCase that counteract the effects of disease-causing variants and restore lysosomal GCase activity in cells.
Read more

Tomás Martins
Tomás and his collaborators engineered a novel CAR T cell that enhances glioblastoma clearance through paracrine immune modulation.
Read more

Submit Your Research
Have you published an article, poster or talk using Promega products? We would love to support you by showcasing your research.
Find out more
We're proud to present the research of Swiss scientists working hard to improve our understanding of life.
Scroll down to read about them, their research, and the Promega technologies they use.

*These authors contributed equally.
FCRL3 is an immunoregulatory receptor that restrains the activation of human memory T lymphocytes
J. Exp. Med. 2026 Jan 5 (epub Oct. 2025); ;223(1):e20242474. doi: 10.1084/jem.20242474. PMID: 41091129; PMCID: PMC12524113
Adequate T cell regulation is indispensable to protect us against pathogens, abnormally proliferating cells, but also autoimmune diseases. A variety of immune cells, including certain T lymphocyte subsets, express the human transmembrane receptor FCRL3. Genetic variants in FCRL3 have been linked to autoimmune conditions such as multiple sclerosis and rheumatoid arthritis, yet its physiological ligands and functions remain poorly defined. The authors of this study uncover a new role for FCRL3 in regulating memory T cell activity.
Read the full publication online: https://rupress.org/jem/article/223/1/e20242474/278373/FCRL3-is-an-immunoregulatory-receptor-that

About Elena and Mehrpouya
Elena and Mehrpouya are currently pursuing their PhD degrees in the Molecular Immunology Laboratory led by Dr. Silvia Monticelli, at the Institute for Research in Biomedicine (IRB) in Bellinzona. Their research focuses on elucidating the mechanisms regulating human T cell responses in inflammatory conditions.

Promega Product Used
Sequencing Grade Modified Trypsin reproducibly generates peptide fragments for mass spectrometry through protein cleavage at lysine and arginine sites. It was engineered to resist autolysis and treated with TPCK to improve specificity, ensuring consistent activity throughout your workflow and reducing background peptides in your spectra.

Ilaria Fregno*, Natalia Pérez-Carmona*, Mikhail Rudinskiy, Tatiana Soldà, Timothy J Bergmann, Ana Ruano, Aida Delgado, Elena Cubero, Manolo Bellotto, Ana María García-Collazo, Maurizio Molinari
*These authors contributed equally.
Allosteric Modulation of GCase Enhances Lysosomal Activity and Reduces ER Stress in GCase-Related Disorders
Int J Mol Sci. 2025 May 6. 26(9):4392. doi: 10.3390/ijms26094392. PMID: 40362629; PMCID: PMC12072338.
Mutations in the GBA1 gene, encoding the glucocerebrosidase (GCase) enzyme, lead to defective GCase protein folding, which impairs its lysosomal delivery and function. GCase variants have been associated with Gaucher disease and constitute an important risk factor for Parkinson's disease and Dementia with Lewy Bodies. Current treatments, while effective, face certain limitations linked to cost, side effects, and failure to boost GCase activity. The authors of this study propose a new therapeutic approach to tackle GCase-related diseases, based on structurally targeted allosteric regulators (STAR).
Variants in the GBA1 gene, encoding the lysosomal enzyme glucosylceramidase beta 1 (GCase), are linked to Parkinson’s disease (PD) and Gaucher disease (GD). Heterozygous variants increase PD risk, while homozygous variants lead to GD, a lysosomal storage disorder. Some GBA1 variants impair GCase maturation in the endoplasmic reticulum, blocking lysosomal transport and causing glucosylceramide accumulation, which disrupts lysosomal function. This study explores therapeutic strategies to address these dysfunctions. Using Site-directed Enzyme Enhancement Therapy (SEE-Tx®), two structurally targeted allosteric regulators (STARs), GT-02287 and GT-02329, were developed and tested in GD patient-derived fibroblasts with relevant GCase variants. Treatment with GT-02287 and GT-02329 improved the folding of mutant GCase, protected the GCase(Leu444Pro) variant from degradation, and facilitated the delivery of active GCase to lysosomes. This enhanced lysosomal function and reduced cellular stress. These findings validate the STARs’ mechanism of action and highlight their therapeutic potential for GCase-related disorders, including GD, PD, and Dementia with Lewy Bodies.
Read the full publication online: https://www.mdpi.com/1422-0067/26/9/4392

About Ilaria and Mikhail
Ilaria and Mikhail obtained their PhDs in cell biology from the ETH Zurich. Currently researchers in the Molinari group at the Institute for Research in Biomedicine (IRB) in Bellinzona, they are committed to understanding the disease determinants in protein misfolding and their role in the pathogenesis of rare disorders.

Promega Product Used
HaloTag® ligands are synthetic molecules that covalently bind to HaloTag® fusion proteins, enabling precise and flexible protein labeling. The Janelia Fluor® HaloTag® ligands offer exceptional brightness, photostability, and cell permeability—ideal for live-cell imaging. With these dyes, you can track protein dynamics with minimal background and with high clarity.

Enhancing anti-EGFRvIII CAR T cell therapy against glioblastoma with a paracrine SIRPγ-derived CD47 blocker
Nat Commun; Nov. 2024; 15(1):9718; doi: 10.1038/s41467-024-54129-w; PMID: 39521782; PCMID: PMC11550474
Glioblastoma (GBM) is an aggressive form of brain cancer that cannot be cured using conventional chemo- and radiotherapy or surgery. While chimeric antigen receptor (CAR) T cell therapy holds great promise for GBM treatment, its efficacy is often hampered by tumor heterogeneity and a strongly immunosuppressive tumor microenvironment. To address the urgent need for more effective treatments, the authors of this study propose an innovative approach that combines targeted CAR T cell therapy with the stimulation and activation of cells within the GBM immune compartment. Read more about their study below and discover how NanoLuc® and Fluorofurimazine (Ffz) helped monitor the efficacy of their approach.
Read the full publication online: https://www.nature.com/articles/s41467-024-54129-w

About Tomás
Tomás completed his PhD degree at the University of Basel, where he explored immune-based therapy approaches and how to harness them to improve outcome in malignant brain tumors. Driven by translational goals, he seeks to bring innovative, patient-centered treatments to clinical practice.

Promega Product Used
Nano-Glo® Fluorofurimazine In Vivo Substrate (FFz) is a substrate designed specifically for in vivo detection of NanoLuc® luciferase and NanoLuc® fusion proteins in animal imaging studies. This optimized, aqueous-soluble detection reagent provides increased substrate bioavailability and bright, stable signals. It also offers flexible delivery options with handling requirements compatible with in vivo workflows.

Bi-allelic variants in COQ8B, a gene involved in the biosynthesis of coenzyme Q10, lead to non-syndromic retinitis pigmentosa
Am J Hum Genet, 2024, Oct 3; 111(10):2299-2306; doi: 10.1016/j.ajhg.2024.08.005; PMID: 39226897
Inherited retinal diseases, such as retinitis pigmentosa (RP), are genetically heterogeneous diseases that cause progressive loss of the retina's photoreceptor cells, eventually leading to blindness. Although more than 100 gene variants have been associated with the pathogenesis of RP, the heritability and molecular mechanisms underlying this disease still present significant knowledge gaps. The authors of this study combined state-of-the-art genomic and protein analysis techniques to investigate the potential causes of retinal damage in RP patients.
Retinitis pigmentosa (RP) is a Mendelian disease characterized by gradual loss of vision, due to the progressive degeneration of retinal cells. Genetically, it is highly heterogeneous, with pathogenic variants identified in more than 100 genes so far. Following a large-scale sequencing screening, we identified five individuals (four families) with recessive and non-syndromic RP, carrying as well bi-allelic DNA changes in COQ8B, a gene involved in the biosynthesis of coenzyme Q10. Specifically, we detected compound heterozygous assortments of five disease-causing variants (c.187C>T [p.Arg63Trp], c.566G>A [p.Trp189Ter], c.1156G>A [p.Asp386Asn], c.1324G>A [p.Val442Met], and c.1560G>A [p.Trp520Ter]), all segregating with disease according to a recessive pattern of inheritance. Cell-based analysis of recombinant proteins deriving from these genotypes, performed by target engagement assays, showed in all cases a significant decrease in ligand-protein interaction compared to the wild type. Our results indicate that variants in COQ8B lead to recessive non-syndromic RP, possibly by impairing the biosynthesis of coenzyme Q10, a key component of oxidative phosphorylation in the mitochondria.
Read the full publication online.

About Ana
Ana currently works as a postdoctoral researcher at the Institute of Molecular and Clinical Ophthalmology Basel (IOB), in the Ophthalmic Genetics group led by Prof. Carlo Rivolta. She applies her expertise in cellular and molecular biology, biochemistry, and microscopy to uncover the causes underlying monogenic and genetically complex ophthalmic diseases. Her aim is to decipher the molecular mechanisms that lead to blindness.
Promega Product Used
The NanoBRET® Target Engagement (TE) Intracellular Kinase Assays are based on the NanoBRET® System, an energy transfer technique designed to measure molecular proximity in living cells. The NanoBRET® TE Assays measure the apparent affinity of test compounds by competitive displacement of the NanoBRET® tracer, reversibly bound to a NanoLuc® luciferase-kinase fusion expressed in cells. In this study, measurement of the affinity of the tracer for wild-type versus mutant proteins was used as a proxy for normal enzymatic activity. Indeed, reduced binding might indicate a compromised or strongly modified catalytic pocket that can lead to functional defects.

Combinatorial Treatment with PARP and MAPK Inhibitors Overcomes Phenotype Switch-Driven Drug Resistance in Advanced Melanoma
Cancer Res., Dec. 2023; 1;83(23):3974-3988. doi: 10.1158/0008-5472.CAN-23-0485; PMID: 37729428
Resistance to targeted therapy constitutes a major challenge in the therapeutic management of melanoma, an aggressive form of skin cancer. The switch from proliferative to highly invasive forms of melanoma that can metastasize often drives the resistance to targeted drugs such as MAPK family inhibitors. Advancing current understanding of phenotype switching at the molecular level offers tremendous potential for developing innovative treatment approaches that will eventually improve the prognosis and treatment of metastatic melanoma.
Metastatic melanoma is either intrinsically resistant or rapidly acquires resistance to targeted therapy treatments, such as MAPK inhibitors (MAPKi). A leading cause of resistance to targeted therapy is a dynamic transition of melanoma cells from a proliferative to a highly invasive state, a phenomenon called phenotype switching. Mechanisms regulating phenotype switching represent potential targets for improving treatment of patients with melanoma. Using a drug screen targeting chromatin regulators in patient-derived three-dimensional MAPKi-resistant melanoma cell cultures, we discovered that PARP inhibitors (PARPi) restore sensitivity to MAPKis, independent of DNA damage repair pathways. Integrated transcriptomic, proteomic, and epigenomic analyses demonstrated that PARPis induce lysosomal autophagic cell death, accompanied by enhanced mitochondrial lipid metabolism that ultimately increases antigen presentation and sensitivity to T-cell cytotoxicity. Moreover, transcriptomic and epigenetic rearrangements induced by PARP inhibition reversed epithelial–mesenchymal transition-like phenotype switching, which redirected melanoma cells toward a proliferative and MAPKi-sensitive state. The combination of PARP and MAPKis synergistically induced cancer cell death both in vitro and in vivo in patient-derived xenograft models. Therefore, this study provides a scientific rationale for treating patients with melanoma with PARPis in combination with MAPKis to abrogate acquired therapy resistance.
Significance: PARP inhibitors can overcome resistance to MAPK inhibitors by activating autophagic cell death and reversing phenotype switching, suggesting that this synergistic combination could help improve the prognosis of patients with melanoma.
Read the full publication online: https://aacrjournals.org/cancerres/article/83/23/3974/730142/Combinatorial-Treatment-with-PARP-and-MAPK

About Lorenza
Lorenza worked as a URPP fellow in translational cancer research and postdoctoral researcher at the University of Zurich (UZH) and University Hospital Zurich (USZ). She is currently exploring opportunities to advance her scientific career. Utilizing her extensive expertise in molecular biology, she applied multidisciplinary approaches to unravel the complexity of melanoma biology. Lorenza is dedicated to pursuing innovative approaches advancing melanoma treatments and aims to make a significant impact on patients' lives.

Promega Product Used
CellTiter-Glo® 3D Cell Viability Assay uses the same reliable chemistry as the classic CellTiter-Glo® Assay and has been optimized to accurately determine cell viability in 3D models. By quantifying ATP levels using a high sensitivity luminescence-based readout, it is an ideal tool to assess drug toxicity in spheroids.

Methylation-based age estimation of different types of bones
Conference abstract. This project was presented at the 44th Spurenworkshop in Frankfurt (March 2024).
Accurate age estimation from forensically relevant traces, for instance using DNA methylation analysis, is essential for supporting forensic casework. While relevant DNA methylation markers and predictive models have been extensively validated for tissues such as blood or saliva, datasets derived from bone samples are comparatively small and existing models remain understudied. To ensure reliable age determination from bones, there is a need for the validation and optimization of available forensic DNA phenotyping models.
Die Altersschätzung mittels DNA-Methylierung aus forensisch relevanten Spuren ist eine derzeit intensiv erforschte Anwendung im Bereich der Forensischen DNA Phänotypisierung. Da sie aktuell in immer mehr Laboren europaweit in die forensische Fallarbeit implementiert wird, ist es wichtig die verschiedenen verfügbaren Modelle gründlich auf ihre Zuverlässigkeit in der Anwendung zu testen.
Wie allgemein bekannt ist, erfordern unterschiedliche Gewebetypen unterschiedliche Marker und Modelle, um die höchstmögliche Schätzgenauigkeit zu erreichen. Ein forensisch relevantes Gewebe, das vergleichsweise weniger Aufmerksamkeit bei DNA-Methylierungsanalysen erhalten hat als beispielsweise Blut oder Speichel, ist Knochengewebe. Obwohl für Knochen bereits Marker erforscht und Modelle für unterschiedliche Technologien (z.B. MPS oder SNaPshot) entwickelt wurden, sind die Datensätze oft vergleichsweise klein, und die verwendete Probensammlung ist nicht immer detailliert beschrieben.
In unserer Studie testen wir ein existierendes MPS-basiertes Altersschätzungsmodell für Knochen an fünf verschiedenen Knochentypen (Rippe, Schlüsselbein, Oberschenkelknochen, Felsenbein und Beckenkamm). Um die Schätzungen der unterschiedlichen Knochen optimal miteinander vergleichen zu können, wurden von jeder obduzierten Person alle fünf Knochentypen gesammelt.
Unsere Fragestellung ist, ob verschiedene Knochentypen in den von uns untersuchten Markern die gleichen Methylierungsmuster aufweisen, und ob wir unterschiedliche Schätzgenauigkeiten für jeden Knochentyp finden. Entsprechend werden wir das geschätzte Alter zwischen den einzelnen Knochentypen vergleichen, um bei Bedarf das Modell bzw. die Auswahl der CpG sites innerhalb der untersuchten Marker anpassen und optimieren zu können.
Age estimation using DNA methylation from forensically relevant traces is an at the moment intensively researched application in the field of Forensic DNA Phenotyping. As it is currently being implemented in forensic casework in an increasing number of laboratories across Europe, it is important to thoroughly test the various available models for their reliability in application.
As generally known, different tissue types require different markers and models to achieve the highest possible age estimation accuracy. One forensically relevant tissue that has received comparatively less attention in DNA methylation analyses than, for example, blood or saliva is bone tissue. Although markers and models for bones have been investigated with various technologies (e.g., MPS or SNaPshot), the datasets are often comparatively small, and the sample collection used is not always described in great detail.
In our study, we are testing an existing MPS-based age estimation model for bones on five different types of bones (rib, clavicle, femur, petrous bone and pelvic crest). To optimally compare the age estimations of these different bones, all five bone types were collected from each autopsied individual.
Our research question is whether different types of bones in the markers we investigated exhibit the same methylation patterns and whether we find different estimation accuracies for each bone type. Accordingly, we will compare the estimated age between the different bone types, in order to adjust and optimize the model or the selection of CpG sites within the investigated markers if necessary.

About Charlotte
Charlotte is currently working as a doctoral researcher at the Institute of Forensic Medicine at the University of Zurich, Switzerland. Using Promega's MethylEdge® Bisulfite Conversion System, she and her collaborators are investigating the relevance of using DNA methylation for age determination in different bone types. Fueled by her passion for forensic genetics, her aim is to advance current understanding of available forensic phenotyping methods and develop their applications for work with difficult or unusual sample types.
Promega Product Used
The MethylEdge® Bisulfite Conversion System provides a rapid, efficient method to perform bisulfite conversion with minimal DNA fragmentation in less than two hours. This all-inclusive kit produces intact, usable, DNA for many downstream applications, including qPCR, arrays, and sequencing. Requiring minimal preparation and hands-on time, it is an ideal companion for streamlined DNA methylation analysis workflows.

*These authors contributed equally.
Engineered reporter phages for detection of Escherichia coli, Enterococcus, and Klebsiella in urine
Nat Commun. 2023 Jul 20;14(1):4336. doi: 10.1038/s41467-023-39863-x. PMID: 37474554; PMCID: PMC10359277
Common bacterial infections often present complex, polymicrobial disease etiology and are therefore challenging to diagnose using standard methods. As a result, broad-range antibiotics are preventively prescribed to treat such infections, contributing to the antimicrobial resistance crisis. More sensitive detection techniques utilizing reporter bacteriophage constitute a promising alternative for the specific and reliable identification of certain types of bacterial pathogens in patients.
The rapid detection and species-level differentiation of bacterial pathogens facilitates antibiotic stewardship and improves disease management. Here, we develop a rapid bacteriophage-based diagnostic assay to detect the most prevalent pathogens causing urinary tract infections: Escherichia coli, Enterococcus spp., and Klebsiella spp. For each uropathogen, two virulent phages were genetically engineered to express a nanoluciferase reporter gene upon host infection. Using 206 patient urine samples, reporter phage-induced bioluminescence was quantified to identify bacteriuria and the assay was benchmarked against conventional urinalysis. Overall, E. coli, Enterococcus spp., and Klebsiella spp. were each detected with high sensitivity (68%, 78%, 87%), specificity (99%, 99%, 99%), and accuracy (90%, 94%, 98%) at a resolution of ≥103 CFU/ml within 5 h. We further demonstrate how bioluminescence in urine can be used to predict phage antibacterial activity, demonstrating the future potential of reporter phages as companion diagnostics that guide patient-phage matching prior to therapeutic phage application.
Read the full publication online: https://www.nature.com/articles/s41467-023-39863-x

About Jiemin
Jiemin is currently working as a doctoral researcher at the Institute of Food, Nutrition, and Health at ETH Zurich, Switzerland. Utilizing the Nanoluciferase reporter system, she and her colleagues have developed a diagnostic tool for the rapid and sensitive detection of bacterial pathogens in patient urine. Collaborating with partners from the University Hospital Zurich and University Hospital Balgrist, she is focused on advancing phage-based therapeutics to address urinary tract infections. Her goal is to integrate her expertise in molecular phage biology and genetic engineering to enhance patient care and combat the growing challenge of antimicrobial resistance.
Promega Product Used
The Nano-Glo® Luciferase Assay System provides a simple, single-addition reagent that generates a stable and long-lasting glow-type signal in the presence of NanoLuc® luciferase. Because of its small size, NanoLuc® luciferase reporter gene can be inserted into viral genomes without disrupting packaging or enzyme activities. This allows for easy monitoring of viral processes, such as infection of bacteria by reporter bacteriophages.

Isoform-specific, Semi-quantitative Determination of Highly Homologous Protein Levels via CRISPR-Cas9-mediated HiBiT Tagging
Bio Protoc. 2023 Jul 20;13(14):e4777. doi: 10.21769/BioProtoc.4777. PMID: 37497448; PMCID: PMC10366989
Genetic redundancy between protein family members is essential to maintain continuous cellular function. However, extensive sequence homology also constitutes a major obstacle in the detection and quantification of distinct protein isoforms. Novel approaches to specifically tag endogenous protein homologs can facilitate their analysis in cells and overcome certain limitations of standard methods relying on isoform-specific antibodies.
Many protein families consist of multiple highly homologous proteins, whether they are encoded by different genes or originating from the same genomic location. Predominance of certain isoforms has been linked to various pathological conditions, such as cancer. Detection and relative quantification of protein isoforms in research are commonly done via immunoblotting, immunohistochemistry, or immunofluorescence, where antibodies against an isoform-specific epitope of particular family members are used. However, isoform-specific antibodies are not always available, making it impossible to decipher isoform-specific protein expression patterns. Here, we describe the insertion of the versatile 11 amino acid HiBiT tag into the genomic location of the protein of interest. This tag was developed and is distributed by Promega (Fitchburg, WI, USA). This protocol describes precise and specific protein expression analysis of highly homologous proteins through expression of the HiBiT tag, enabling protein expression quantification when specific antibodies are missing. Protein expression can be analyzed through traditional methods such as western blotting or immunofluorescence, and also in a luciferase binary reporter system, allowing for reliable and fast relative expression quantification using a plate reader.
Read the full publication online: https://bio-protocol.org/en/bpdetail?id=4777&type=0

About Kristina
Kristina is currently working as a postdoctoral research scientist at the University of Utah in Salt Lake City (USA). She completed her MD-PhD studies at the University of Bern (CH) and Scripps Research La Jolla (USA), where she focused on the role of hexokinase 3 (HK3) in the development of acute myeloid leukemia (AML). Using the HiBiT protein tagging system, she and her colleagues were able to discover a novel role for HK3 in mediating cell death during the treatment of AML cells. In her current role, Kristina is putting her expertise in cellular energy metabolism to use and researching how mitochondrial energy production is regulated in health and disease.
Promega Product Used
The HiBiT protein tagging system allows for simple and sensitive bioluminescent protein quantitation. The 11 amino acid HiBiT tag can be added easily to a protein of interest using CRISPR-mediated genome editing. The total amount of HiBiT-tagged protein can be evaluated in live cells or cellular lysates through the reconstitution of NanoBiT® enzyme in the presence of LgBiT protein. In addition to this direct detection method, HiBiT-tagged proteins can also be detected via immunostaining using the anti-HiBiT monoclonal antibody.

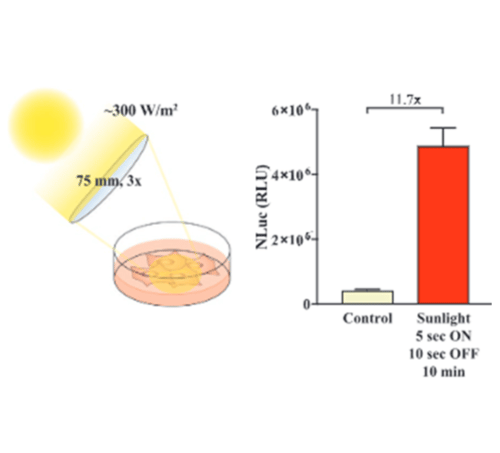
Sunlight-Controllable Biopharmaceutical Production for Remote Emergency Supply of Directly Injectable Therapeutic Proteins
Read the full paper online: https://onlinelibrary.wiley.com/doi/10.1002/smll.202202566

About Adrian
During his studies in Molecular Health Science, Adrian Stefanov developed an interest in personalized medicine and unconventional therapeutic approaches. This inspired him for a PhD at ETH Zurich where he applied specially modified human cells for diabetes therapy. He established a gene switch that enables electronic devices to be interfaced with the engineered cells for a digital fine-tuning of the insulin amounts released from the cells into the body. In addition to that, he developed a manufacturing strategy for ready-to-use therapeutic protein solutions at remote locations, as described in the article above.
Promega Product Used
The Nano-Glo® Luciferase Assay System provides a simple, single-addition reagent that generates a glow-type signal in the presence of NanoLuc® luciferase with a half-life of approximately 120 minutes in commonly used tissue culture media.

Circulating tumor DNA for comprehensive noninvasive monitoring of lymphoma treated with ibrutinib plus nivolumab
To advance the use of circulating tumor DNA (ctDNA) applications, their broad clinical validity must be tested in different treatment settings, including targeted therapies.
To advance the use of circulating tumor DNA (ctDNA) applications, their broad clinical validity must be tested in different treatment settings, including targeted therapies.
Using the prespecified longitudinal systematic collection of plasma samples in the phase 1/2a LYM1002 trial (registered on www.clinicaltrials.gov as NCT02329847), we tested the clinical validity of ctDNA for baseline mutation profiling, residual tumor load quantification, and acquisition of resistance mutations in patients with lymphoma treated with ibrutinib+nivolumab. Inclusion criterion for this ancillary biological study was the availability of blood collected at baseline and cycle 3, day 1.
Overall, 172 ctDNA samples from 67 patients were analyzed by the LyV4.0 ctDNA Cancer Personalized Profiling Deep Sequencing Assay. Among baseline variants in ctDNA, only TP53 mutations (detected in 25.4% of patients) were associated with shorter progression-free survival; clones harboring baseline TP53 mutations did not disappear during treatment. Molecular response, defined as a >2-log reduction in ctDNA levels after 2 cycles of therapy (28 days), was achieved in 28.6% of patients with relapsed diffuse large B-cell lymphoma who had ≥1 baseline variant and was associated with best response and improved progression-free survival.
Clonal evolution occurred frequently during treatment, and 10.3% new mutations were identified after 2 treatment cycles in nonresponders. PLCG2 was the topmost among genes that acquired new mutations. No patients acquired the C481S BTK mutation implicated in resistance to ibrutinib in CLL.
Collectively, our results provide the proof of concept that ctDNA is useful for noninvasive monitoring of lymphoma treated with targeted agents in the clinical trial setting.
Read the full publication online: https://ashpublications.org/bloodadvances/article/5/22/4674/476850/Circulating-tumor-DNA-for-comprehensive

About Alessio
Since 2016, Alessio Bruscaggin has worked as a senior Post-doc in the Laboratory of Experimental Hematology of the Institute of Oncology Research in Bellinzona, Switzerland, where he coordinates next-generation sequencing projects in lymphoproliferative disorders.
He has a large experience in mature B-cell tumor genetics and in the development of ultra-deep next-generation sequencing pipelines, which will represent the methodological platform of all the mutation analysis. He actively participated in several projects aiming at the characterization of the molecular pathogenesis of B-cell tumors and at the translation of biological information into markers for disease diagnosis, prognostication, and treatment. He also contributed to the characterization of the splenic marginal zone lymphoma and CLL genomes, and to the development of plasma cell-free DNA as a tool to inform on tumor genetics in lymphomas.

Promega Product Used
Maxwell®️ instruments are used together with cartridge-based purification kits to quickly extract DNA or RNA from many different sample types.
The automated purification process consistently delivers quality nucleic acid, freeing up scientists’ time for other important work. The Maxwell RSC LV cfDNA kit purifies circulating cell-free DNA from up to 8ml or more of plasma.

Nikolai Püllen,a Hailey Gahlon,a Shana J. Sturlaa*
Removal of Common Alterations in Genomic DNA for Accurate Damage Sequencing
The structural integrity of DNA is constantly altered by exposure to metabolic byproducts, chemicals or radiation. Persisting DNA damage can result in mutations and genome instability. To better understand biological influences of DNA damage formation, new DNA damage sequencing methods are rapidly emerging.
*e-mail: sturlas@ethz.ch
aDept. of Health Sciences and Technology, ETH Zurich, Schmelzbergstr. 9, 8092 Zurich,Switzerland;
Acknowledgment: We acknowledge funding from the Swiss National Science Foundation.
References
(1) Mingard, C. et al. Chem. Soc. Rev. 2020, 49 (20), 7354–7377.

About Nikolai
Nikolai Püllen graduated from the University of Münster, Germany, with a Master in Chemistry. He joined Johnson and Johnson R&D in Zurich, as an industrial research intern. Currently, he is a Ph.D. student at the ETH in Zurich working on DNA damage sequencing in the Laboratory of Toxicology. Nikolai enjoys research as part of understanding how life works and as a mean to engage in scientific communication. He is curious, creative and also a project management enthusiast, which helps when conducting research.
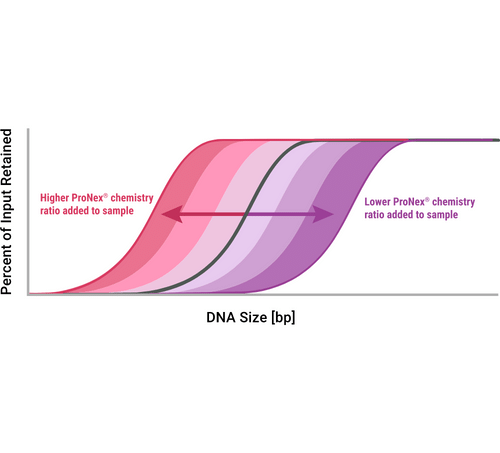
Promega Product Used
The ProNex® Size-Selective Purification System enables the rapid and efficient magnetic resin-based purification of double-stranded DNA (dsDNA) for next-generation sequencing (NGS), polymerase chain reaction (PCR) and general molecular biology applications.

Paraquat-induced cholesterol biosynthesis proteins dysregulation in human brain microvascular endothelial cells
About Tatjana
Tatjana Vujić earned her Master's Degree in Pharmaceutical Sciences at the University of Geneva in Switzerland. During her Master's Degree, she joined the Nestlé Research Center R&D in the Vitamins and Phytonutrients Department in Lausanne. Currently doing her Ph.D. thesis in the Translational Biomarker Group at the University of Geneva, she studies the xenobiotic effects on human brain endothelial cells based on protein modulations, including cholesterol proteins. Persistent, curious, and optimistic by nature, Tatjana enjoys facing and meet the challenges of scientific discovery.
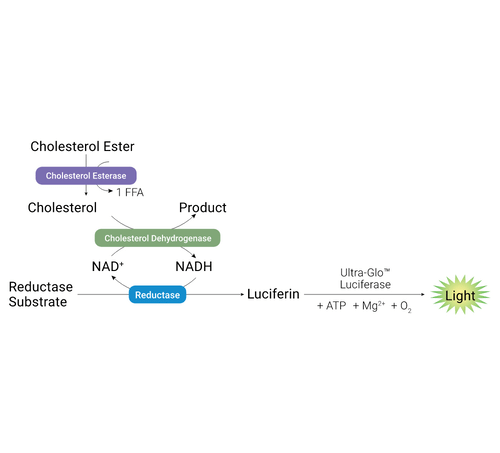
Promega Product Used
The Cholesterol/Cholesterol Ester-Glo™ Assay is a bioluminescent assay for rapid and sensitive measuring of cholesterol and cholesterol esters in cultured cell lysates and other biological samples, such as lipoprotein fractions, cell culture medium, serum and tissue homogenates. Cholesterol is an essential lipid involved in steroidogenesis, bile acid synthesis, cell signaling and maintenance of membrane structure.
Submit Your Research
Have you published an article recently using Promega products? Will you present a talk or a poster at a conference soon? We would love to support you by showcasing your research on our website and social media and offering you additional rewards.
Get in touch with Anna to find out more.
This spotlight program is opened to participants conducting their research in Switzerland.
A participant can apply with a talk or a poster only once. The participant must provide proof of registration at a conference and gives Promega the right to showcase their research on Promega.com and Promega’s social media accounts. Promega is not responsible for the validity and origin of the work published. Promega reserves the right to exclude persons or organizations from participating in this program.
By taking part in the Spotlight program, the participant agrees that Promega AG will save the necessary data for the purpose of the program and for the duration of the program and beyond. Participants have the right to revoke their consent at any time, thus canceling their partaking in the Spotlight program.
Promega AG reserves the right, in its sole discretion, to cancel, terminate, modify or suspend the promotion at any time of the program duration.
