Lining Up the Scripts: Reverse Transcriptase Comparison Study
Manuel Ammerschläger1, Petra Kioschis1, Eric Vincent2, Jeff Briganti2 and Trista Schagat2
1Hochschule Mannheim, University of Applied Sciences, and 2Promega Corporation
Publication Date: 2009
Abstract
Accurate gene expression analysis depends on the reverse transcriptase used as well as other factors (e.g., RNA quality and detection method). The ideal reverse transcriptase will robustly reverse transcribe from the priming site to the 5´ end of RNAs. We performed a direct functional comparison of popular reverse transcriptases to determine which offers superior performance. The new Promega GoScript™ Reverse Transcriptase performed as well as or better than Invitrogen’s SuperScript® II and SuperScript® III and Qiagen’s Omniscript® and Sensiscript® reverse transcriptases in terms of sensitivity of transcript detection, transcript length and sensitivity to ethanol contamination. We further compared GoScript™ and SuperScript® III reverse transcriptases using a difficult transcript and the standard elevated reaction temperature recommended for SuperScript® III (50°C). GoScript™ Reverse Transcriptase outperformed SuperScript® III. These data support the use of GoScript™ Reverse Transcriptase as a high-quality reverse transcriptase for gene expression analysis.
Introduction
Reverse transcriptases (RTs), also known as RNA-directed DNA polymerases, were first identified in retroviruses(1) (2) . In addition to the critical role these enzymes play in a variety of major human diseases (HIV, hepatitis and some cancers), RTs are a fundamental component of the molecular biologist’s toolbox. RT activity is critical for many basic techniques including real-time and endpoint RT-PCR, labeled-probe generation for microarrays and cDNA cloning. The ideal reverse transcriptase is robust (highly active under a variety of conditions) and converts all primed RNA within a sample to cDNA, regardless of its abundance, length or secondary structure.
The most characterized RTs used for molecular biology are the retroviral RTs: avian myeloblastosis virus (AMV) and Moloney murine leukemia virus (MMLV). Genetic engineering and development of proprietary RT-enhancing buffers have led to the commercial availability of new enzymes that offer superior performance over these naturally occurring RTs.
We performed a head-to-head comparison of five popular trademarked RTs: GoScript™ (Promega Cat.# A5003), SuperScript® II and SuperScript® III (Invitrogen Cat.# 18064 and 18080, respectively), and Omniscript® and Sensiscript® (Qiagen Cat.# 205113 and 205213, respectively) reverse transcriptases. We compared these five RTs for their utility in detecting trace transcripts by real-time PCR, ability to transcribe a long RNA and sensitivity to ethanol, a common contaminant in RNA preparations. We also compared SuperScript® III and GoScript® reverse transcriptases for their ability to transcribe a challenging GC-rich mRNA.
"In all cases, GoScript™ Reverse Transcriptase performed as well as or better than all other reverse transcriptases tested."
Transcript Sensitivity
Gene expression analysis is the most common use for RTs. The ideal RT will reverse transcribe even the least abundant mRNAs within a pool, allowing single-cell transcript detection. (Note: The PCR conditions used also will influence detection sensitivity.) To compare RT sensitivities of transcript detection, we used each enzyme and oligo(dT) as the primer to reverse transcribe serial dilutions of mouse liver total RNA representing 1 to 10,000 cell equivalents (assuming 20pg of total RNA per cell). Two microliters of each cDNA synthesis reaction then was analyzed using TaqMan® real-time PCR (Applied Biosystems). The following four transcripts were analyzed (listed from highest to lowest abundance): glyceraldehyde-3-phosphate dehydrogenase (GAPDH), lamin A, transformation-related protein 53 (trp53) and cyclin-dependent kinase 9 (cdk9).
All RTs allowed detection down to a single cell of the high-abundance transcript GAPDH (Figure 1, Panel A). Only GoScript™ Reverse Transcriptase allowed linear detection of lamin A down to a single cell (Figure 1, Panel B). For the lower-abundance trp53 and cdk9 transcripts, detection was lost for all RTs at 10 and 1,000 cells, respectively, regardless of which enzyme was used (Figure 1, Panels C and D). Based on these results, we concluded there is no dramatic distinction in the utility of these RTs with regard to sensitivity of transcript detection in reverse transcription-quantitative PCR(3) .
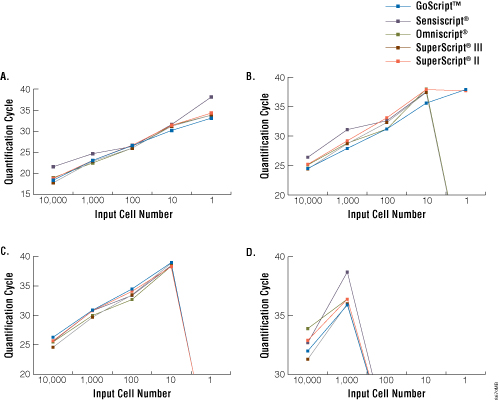
Figure 1. Sensitivity of detection of GAPDH, lamin A, Trp53 and Cdk9 transcripts. Serial dilutions of mouse liver total RNA (Clontech Laboratories, Inc.) at 1–10,000 cell equivalents were reverse transcribed using random primers according to the manufacturer's instructions for each RT. cDNA then was analyzed for four transcripts: high-abundance GAPDH (Panel A), moderate-abundance lamin A (Panel B) and trp53 (Panel C), and low-abundance cdk9 (Panel D) by TaqMan® real-time PCR (Applied Biosystems assays Mm99999915_g1, Mm00497783_m1, Mm00441964_g1 and Mm00499939_g1, respectively; 2µl of cDNA per 25µl reaction). Data are the average quantification cycle (Cq) for two separate cDNA reactions analyzed in duplicate. Samples with no detectable amplification were assigned a quantification cycle of zero (i.e., lines intersect the X axis).
Note: Cq is often referred to as cycle threshold (CT).
Transcript Length
Reverse transcription of full-length mRNAs is critical for cloning or when the PCR target is at the 5´ end of a transcript that will be oligo(dT)-primed. Without efficient extension during the RT reaction, portions of the transcripts may be lost to further analysis. We tested the ability of each RT to reverse transcribe an 8.9kb adenomatosis polyposis coli (APC) transcript from human total RNA. All reactions used standard incubation times of 50–60 minutes. We compared an oligo(dT)-primed reaction to a random-primed control reaction. cDNA was analyzed by endpoint PCR using primers that amplify a 940bp region at the 5´ end (forward primer included ATG start codon).
GoScript™ Reverse Transcriptase gave more PCR product for both the random-primed and oligo(dT)-primed cDNA than any other RT (Figure 2). All RTs yielded a product from the random-primed reaction, but when oligo(dT) was used as the primer, little to no product was detected for Omniscript® and Sensiscript® RTs,indicating the RTs did not efficiently reach the 5´ end of the transcript. SuperScript® II and SuperScript® III RTs both gave full-length product, but as with the random-primed samples, the product was not as abundant as that from RNA reverse transcribed with GoScript™ Reverse Transcriptase. GoScript™ Reverse Transcriptase was the most effective at reverse transcribing the full-length 8.9kb APC transcript.
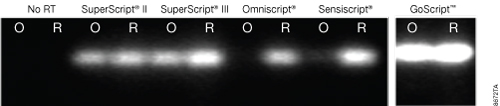
Figure 2. Reverse transcription of an 8.9kb APC transcript. Human colon total RNA (Clontech Laboratories, Inc.) was reverse transcribed using oligo(dT) (O) or random primers (R) according to the manufacturer’s instructions for each reverse transcriptase. cDNA then was analyzed by endpoint PCR using GoTaq® Green Master Mix (Cat.# M7122) with primers for a 940bp amplicon at the 5´ end of the transcript. Each reaction (5µl) was analyzed by agarose gel electrophoresis and ethidium bromide staining. All samples shown were analyzed in the same data set and gel with the same scan settings. Gel images are representative of results from three separate experiments.
Ethanol Sensitivity
Ethanol is routinely used in nucleic acid purification protocols. Insufficient drying of either the RNA pellet or purification column (depending on the method used) can lead to residual ethanol in the final RNA. We examined the ability of an ethanol contaminant to affect each RT. An in vitro-transcribed RNA (1.2kb Kanamycin Positive Control RNA) was reverse transcribed in the presence of 5%, 10% or 20% ethanol, and the results were compared to those of an untreated control. cDNA then was analyzed by real-time PCR using GoTaq® qPCR Master Mix. The relative change in transcript level was determined based on the difference in quantification cycle values in the presence and absence of inhibitor [2(Cqinhibitor–Cqnone)]. All RTs exhibited minimal or no effect at 5% ethanol (Figure 3). At 20% ethanol, all RTs were affected to some extent. GoScript™ Reverse Transcriptase showed the smallest decrease in the ability to detect transcript in the presence of ethanol: 1.07-fold (<10%) change 1.07-fold (<10%) change at 10% ethanol and 1.2-fold (20%) change at 20% ethanol.
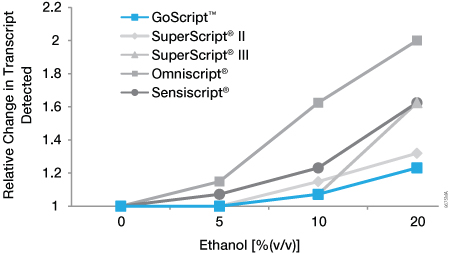
Figure 3. Sensitivity of reverse transcriptases to ethanol inhibition. An in vitro-transcribed RNA (1.2kb Kanamycin Positive Control RNA, Cat.# C1381) was reverse transcribed using oligo(dT) primer according to the manufacturer’s instructions for each RT. Reverse transcription was carried out in the absence and presence 5%, 10% and 20% ethanol. cDNA was analyzed by real-time PCR using GoTaq® qPCR Master Mix (Cat.# A6001). The change in quantification cycle values in the absence and presence of ethanol was determined and converted to relative change in transcript level[2(Cqinhibitor–Cqnone)]. Data are the average of three separate experiments.
Difficult Transcript
Highly structured or GC-rich transcripts can be difficult to reverse transcribe due to premature termination, resulting in truncated cDNAs. The use of elevated temperatures during the RT reaction can help denature the RNA, allowing the RT to advance more effectively. SuperScript® III RT is relatively thermostable and has a standard reaction temperature of 50°C. This is 8°C higher than that of SuperScript® II and GoScript™ reverse transcriptases (42°C) and 13°C higher than that recommended for Omniscript® or Sensiscript® RTs (37°C).
We compared the ability of GoScript™ and SuperScript® III reverse transcriptases to reverse transcribe a challenging transcript, nocturnin (GenBank Accession# NM_012118), which has several regions at >60% GC. GoScript™ reactions were performed at 50°C (as recommended for SuperScript® III) or 42°C (standard GoScript™ reaction condition) prior to endpoint and real-time PCR analyses, respectively. Oligo(dT)-primed cDNAs were analyzed using gene-specific primers for nocturnin and β-2 macroglobulin (B2M), which was amplified as a control.
Gel analysis of the endpoint PCR products showed GoScript™ Reverse Transcriptase was more efficient at producing a nocturnin PCR product (Figure 4, Panel A). Both enzymes were equally efficient at producing the control β-2 macroglobulin product, suggesting GoScript™ Reverse Transcriptase was better able to work through the nocturnin transcript. These results were confirmed by real-time PCR using GoTaq® qPCR Master Mix (Figure 4, Panel B). The difference in quantification cycle values for nocturnin cDNA reverse transcribed with GoScript™ Reverse Transcriptase and SuperScript® III was 2.3 cycles, while there was no significant difference in β-2 macroglobulin detection. This represents a 4.9-fold better detection of the challenging nocturnin transcript with GoScript™ Reverse Transcriptase versus SuperScript® III.
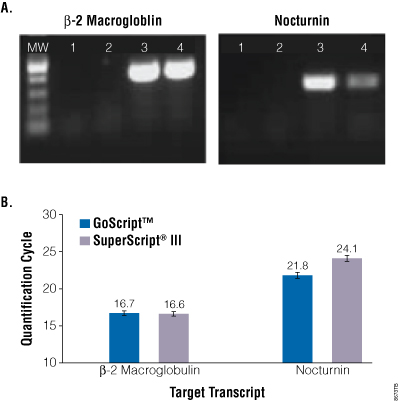
Figure 4. Reverse transcription of a challenging GC-rich transcript. Panel A. Human total RNA was isolated from HEK293 cells using TRI Reagent® (Ambion Cat.# AM9738) and reverse transcribed using oligo(dT) primer and GoScript™ or SuperScript® III reverse transcriptase, following the manufacturers’ instructions with the exception that all reactions were carried out at 50°C. cDNA was analyzed by amplification of β2 macroglobulin (control) or GC-rich nocturnin using GoTaq® Green Master Mix. Reactions were analyzed by agarose gel electrophoresis and ethidium bromide staining. Lane 1. No-template control. Lane 2. No-reverse transcriptase control. Lane 3. GoScript™ Reverse Transcriptase reactions. Lane 4. SuperScript® III reactions. Panel B. Human total RNA was reverse transcribed but at each enzyme’s recommended reaction temperature (GoScript™ Reverse Transcriptase at 42°C and SuperScript® III at 50°C). cDNA was analyzed by real-time PCR for both transcripts using GoTaq® qPCR Master Mix. Data are the average of three independent experiments.
Summary
In addition, GoScript™ Reverse Transcriptase was further tested against SuperScript® III for the ability to reverse transcribe a challenging transcript at elevated temperature. In all cases, GoScript™ Reverse Transcriptase performed as well as or better than all RTs tested. These data demonstrate the high quality and robustness of the GoScript™ Reverse Transcriptase and support its use for highly sensitive gene expression analysis.
References
- Temin, H.M. and Mizutani, S. (1970) RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature 226, 1211–3.
- Baltimore, D. (1970) RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature 226, 1209–11.
- Bustin, S.A. et al. (2009) The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–22.
We compared GoScript™ Reverse Transcriptase to Invitrogen SuperScript® II and SuperScript® III as well as Qiagen OmniScript® and Sensiscript® reverse transcriptases. Several key performance variables were tested: sensitivity of transcript detection, transcript length and sensitivity to a common RNA contaminant, ethanol.