Introducing GoTaq® qPCR Master Mix: The Bright Choice for Dye-Based qPCR
Karen L. Reece, Katharine Driftmier Miller Hoffmann, Cesear Corona, Thomas A. Kirkland, H. Tetsuo Uyeda, Stephen J. Dwight, Mark G. McDougall and Douglas R. Storts
Promega Corporation, Madison, WI, Promega Biosciences, LLC., San Luis Obispo, CA
Publication Date: 2009
Abstract
The new GoTaq® qPCR Master Mix is a 2X master mix for quantitative PCR (qPCR) and features GoTaq® Hot Start Polymerase, a full-length Taq DNA polymerase bound to a proprietary antibody that prevents polymerase activity at room temperature. The master mix is composed of an optimized buffer formulation complete with dNTPs and MgCl2 and includes a new, proprietary dsDNA-binding dye that can be used at a higher concentration than SYBR® Green I because it is less inhibitory in an amplification reaction. The dye concentration in the master mix is optimized to produce significantly brighter fluorescence during qPCR than master mixes containing SYBR® Green I. Excitation and emission of the dye are similar to those of SYBR® Green I, so it is compatible with commonly available real-time PCR instrumentation.
Introduction
Real-time quantitative PCR is a powerful tool to detect and quantify nucleic acids. By incorporating fluorescently labeled probes or nucleic acids or fluorescent double-stranded DNA (dsDNA)-binding dyes into the PCR, product formation can be monitored following each PCR cycle. There are many advantages to using real-time PCR over endpoint PCR. Use of fluorescent dyes to monitor amplification eliminates the need for gel electrophoresis, saving the user time and reducing the potential for contamination. Since product formation can be detected in the exponential phase of PCR where the reaction is most efficient, quantification of starting material is more accurate and sensitive (1). The sensitivity offered by fluorescent dyes also allows detection over a much broader dynamic range than endpoint PCR.
Probe-based chemistries such as TaqMan® technology are advantageous in that the labeled probe, which is specific to the amplicon, produces a fluorescent signal during the enzymatic processing of the probe:amplicon hybrid, indicating the specific synthesis of amplicon. However, these assays are difficult to design and more expensive than dye-based chemistries. dsDNA-binding dyes, such as SYBR® Green I, detect all dsDNA formed during amplification; however, these assays are simple to design and cost-effective. The ability to perform melt curve analysis, which is not possible with the TaqMan® assay, provides a quality control step to verify amplification of the desired amplicon.
GoTaq® qPCR Master Mix (Cat.# A6001) introduces a new, proprietary dsDNA-binding dye, Bryt™ Green dye, that can be used at a higher concentration than SYBR® Green I because it is less inhibitory in an amplification reaction. The dye concentration in the master mix is optimized to produce significantly brighter fluorescence during qPCR than master mixes containing SYBR® Green I. Excitation and emission of the dye are similar to those of SYBR® Green I (Figure 1), so it is compatible with commonly available instrumentation platforms. GoTaq® qPCR Master Mix is a 2X master mix composed of an optimized buffer formulation complete with dNTPs and MgCl2 and features GoTaq® Hot Start Polymerase . GoTaq® Hot Start Polymerase contains full-length Taq DNA polymerase bound to a proprietary antibody that prevents polymerase activity at room temperature. Thermal activation is achieved by incubating the assembled reaction at 95°C for 2 minutes. The proprietary polymerase/buffer formulation accommodates extended cycle numbers (45–50 cycles) and is compatible with thermal cycling programs that include extended activation (95°C for 10 minutes). The master mix comes premixed with a low level of CXR reference dye, which is identical to ROX™ reference dye. The master mix is compatible with existing experimental designs and requires addition of only DNA template, target-specific primers and water.
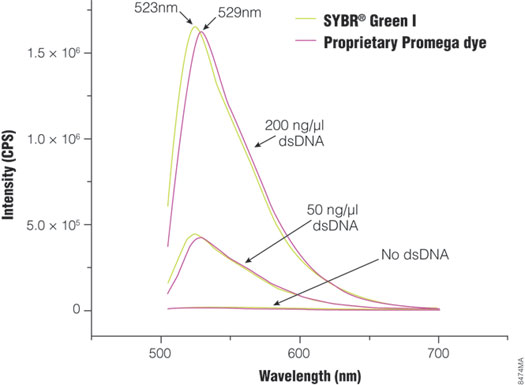
Figure 1. Fluorescence spectra of SYBR® Green I and the proprietary Promega dye in the GoTaq® qPCR Master Mix. Fluorescence emission spectra were collected for both SYBR® Green I and Promega Bryt™ Green in the absence of dsDNA and in the presence of 50ng/µl or 200ng/µl dsDNA. Peak emission wavelength is 523nm for SYBR® Green I and 529nm for Bryt™ Green.
"GoTaq® qPCR Master Mix introduces a new, proprietary dsDNA-binding dye that can be used at a higher concentration than SYBR® Green I because it is less inhibitory in an amplification reaction, producing significantly brighter fluorescence. Excitation and emission of the dye are similar to those of SYBR® Green I."
Methods
Unless otherwise noted, 50μl reactions were prepared, and PCR was performed using an Applied Biosystems 7500 Real Time PCR instrument in 9600 emulation mode and calibrated according to the manufacturer’s instructions using the Pure Dye Calibration Kit (Applied Biosystems). Unless otherwise noted, cycling conditions were as follows: Activation at 95°C for 2 minutes, 40 cycles of denaturation at 95°C for 15 seconds and annealing/extension at 60°C for 60 seconds, followed by melt analysis ramping at 60°C to 95°C.
Results
Brightness and Sensitivity
The brighter fluorescence signal provided by the GoTaq® qPCR Master Mix enables earlier detection of amplification products, resulting in earlier quantification cycle (Cq) values for many targets compared to SYBR® Green I (Figure 2). This property is demonstrated by amplifying glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from serial dilutions of Human Genomic DNA (Cat.# G3041) using either GoTaq® qPCR Master Mix or Company A’s master mix on the same 96-well plate. The instrument was programmed for an initial 10-minute enzyme activation step at 95°C to accommodate the requirements of Company A’s master mix. The significantly brighter fluorescent signal generated with GoTaq® qPCR Master Mix enables the analysis software to call the Cq values up to three cycles earlier.
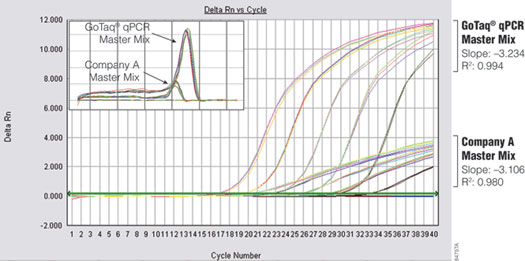
Figure 2. Performance comparison of GoTaq® qPCR Master Mix and Company A's master mix. GAPDH was amplified from tenfold serial dilutions (0.01–100ng) of human genomic DNA. Inset shows comparison of dissociation profiles for both master mixes.
Amplification of a kanamycin phosphotransferase target sequence from plasmid DNA (Figure 3) shows efficient, linear amplification over 8 orders of magnitude with accurate quantification across the entire dynamic range. Alpha V was amplified from serial dilutions of cDNA prepared from Total RNA Positive Control (Part# C199A) and ImProm-II™ Reverse Transcriptase (Cat.# A3802). Figure 4 shows specific and sensitive detection of one copy per reaction.
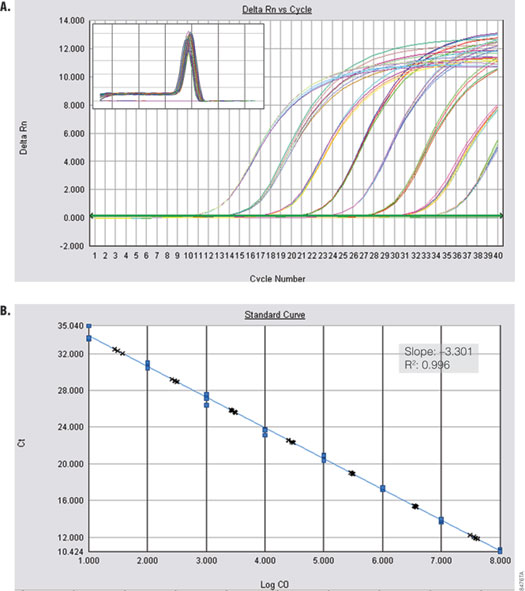
Figure 3. Amplification of kanamycin phosphotransferase from plasmid DNA. Panel A. Amplification curves are shown for tenfold dilutions of 10 to 1 × 108 copies of plasmid. Inset shows the dissociation profile. Panel B. The blue dots in the standard curve represent the Cq values for the tenfold dilution series. The black crosses show quantification of tenfold dilutions of 30 to 3 × 107 copies of plasmid DNA. Inset shows the standard curve data
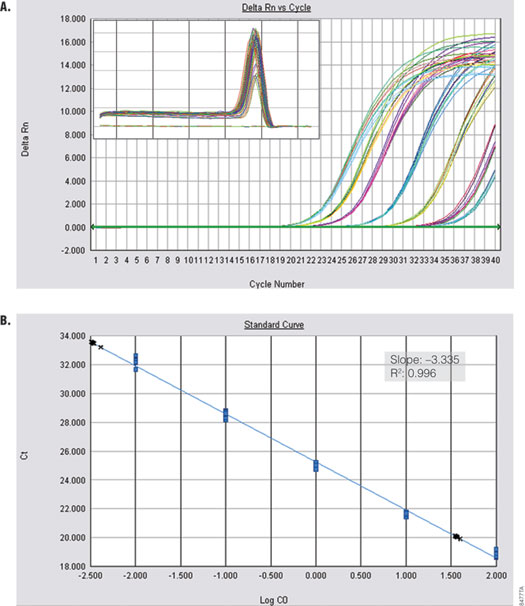
Figure 4. Amplification of Alpha V from cDNA. Alpha V was amplified from serial dilutions of cDNA (0.01–100ng) using the GoTaq® qPCR Master Mix. Panel A. Amplification curves are shown for reactions containing 3pg and 33ng cDNA. Inset shows dissociation profile. Panel B. The standard curve is shown. Blue dots represent tenfold dilutions. Black crosses show accurate quantification of 3pg and 33ng cDNA. Inset shows standard curve data.
Versatility
GoTaq® qPCR Master Mix is compatible with several real-time PCR instrument platforms using preset filters for SYBR® Green I or FAM™ and ROX™ for the passive reference. Depending on instrument detection settings, however, some instruments require a higher concentration of reference dye in the reaction mix. A tube of 100X CXR Reference Dye is provided with the master mix. Below we list real-time PCR instruments tested and indicate which instruments require additional reference dye.
Compatibility of GoTaq® qPCR Master Mix With Various Real-Time PCR Instruments
Instruments that do not require addition of 100X CXR Reference Dye:
- Applied Biosystems 7500 and 7500 Fast Real Time PCR Systems
- Stratagene Mx3005P® Quantitative PCR Systems
- Roche LightCycler® 480
- BioRad Chromo4™ Real-Time Detector
- Eppendorf Mastercycler® ep realplex4 and realplex4 S
Instruments that require addition of 100X CXR Reference Dye to 1X per reaction:
- Applied Biosystems 7000 Sequence Detection System
- Applied Biosystems 7300 Real-Time PCR System
- Applied Biosystems 7700 Sequence Detection System
- Applied Biosystems 7900HT Real-Time PCR System
- Applied Biosystems StepOne® and StepOne® Plus Real-Time PCR Systems
Bio-Rad real-time instruments, such as the iCycler®, iQ™5 and MyCycler™, perform normalization by collecting Dynamic Well Factors using the background signal detected during the first few PCR cycles. Because the background using SYBR® Green I is not sufficient for the instrument to make these calculations, Bio-Rad protocols require the addition of 10nM fluorescein per reaction (3) . GoTaq® qPCR Master Mix was tested on a Bio-Rad iQ™5 instrument with and without addition of fluorescein. Figure 5 shows that amplification of GAPDH from serial dilutions of human genomic DNA was linear and efficient both with and without addition of fluorescein to the master mix. This suggests that the brightness of the proprietary Promega dye provides sufficient signal for the instrument to collect Dynamic Well Factors without addition of fluorescein. When using a Bio-Rad instrument with GoTaq® qPCR Master Mix, users should run a test reaction using their experimental target to determine if it is necessary to add fluorescein to the master mix.
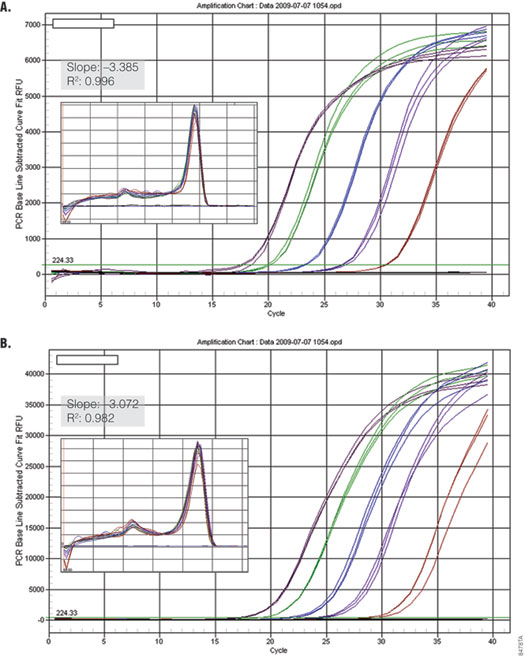
Figure 5. Performance of GoTaq® qPCR Master Mix using the Bio-Rad iQ™5 with and without fluorescein. GAPDH was amplified from serial dilutions of human genomic DNA (0.01–100ng) using the default cycling protocol on the Bio-Rad iQ™ 5 Real Time instrument. Panel A. Detection of GAPDH without fluorescein. Panel B. Detection with 10nM fluorescein added (Bio-Rad Cat.# 170-8780). Insets show standard curve data and dissociation profile.
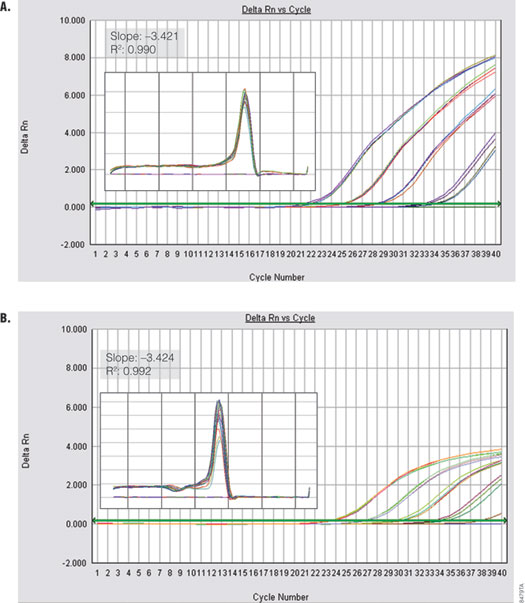
Figure 6. Performance of GoTaq® qPCR Master Mix using a fast cycling protocol. GAPDH was amplified from serial dilutions of human genomic DNA (0.01–10 ng) using the default fast cycling protocol on the Applied Biosystems 7500 FAST Real Time instrument Panel A. GoTaq® qPCR Master Mix. Panel B. Company A’s fast master mix. Insets show standard curve data and dissociation profile.
GoTaq® qPCR Master Mix provides superior amplification and fluorescence qualities in a convenient 2X formulation, which includes a new, proprietary dsDNA-binding dye that results in a significantly brighter signal during qPCR than SYBR® Green I. Since the spectral properties of this dye are similar to SYBR® Green I, the master mix can be easily substituted for other dye-based
Summary
qPCR master mixes. The convenient 2X master mix requires addition of only experimental template DNA, primers and water. It is compatible with common thermal cycling protocols and fast cycling protocols and can be used on any real-time instrument capable of detecting SYBR® Green I or FAM™ dye.
References
- Sundquist, T. (2005) Your key to real-time quantitative PCR. Promega eNotes Web page.
- Bustin, S.A. et al. (2009) The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–22.
- BioRad iCycler iQ™ Real-Time PCR Detection System Instruction Manual, 4006200 Rev E.